Superresolution-actin-and-mitochondria-analysis
Measure granularity spectrum
Description
Granularity spectrum measurement in cells based on finding cells using Cellpose segmentation, and then using a set of Fourier domain band pass filters to amplify various detail levels of the image. The granularity spectrum is built from intensity readouts from the filtered images starting at the lowest frequency image components and then stepwise increasing the cut on and cut of frequencies of the filter. The number of frequency bins is coded in the pipeline structure. This pipeline was used as an alternative to granularity spectrum measurement with Cell Profiler.
- The input of the pipeline is a two-channel image of cells with the channel of interest for spectral measurement, and a second channel with a nuclear stain. Alternatively use an image name “Cells” and “Perform Cellpose segmentation”=No to skip segmentation and directly use cell segments, e.g. during repeated execution of the pipeline with different parameters. No nucleus image is needed this case. The input may be an image series, and in this case the analysis will run on each frame independently. This is the fastest way of processing many images.
- The output is an Excel worksheet where spectral bins are in columns and frames are in rows.
Parameters
| # | Name | Type | Description |
|---|---|---|---|
| 0 | Channel number to analyze (actin) | Integer | Granularity spectrum in this channel will be measured. This channel also serves to identify cells with segmentation |
| 1 | Channel number for nuclei | Integer | Nuclei will help image segmentation with Cellpose. |
| 2 | Perform Cellpose segmentation | Bool | Set this to No if the segmented image named as "Cells" already exists. |
| 3 | Bandwidth | Real | Band width of band pass Butterworth filter in frequency domain pixels |
| 4 | Filter normalization | Text | Normalizes the integral of the 1D or 2D filter function for filtering in Fourier Domain |
| 5 | Order | Real | Order of the cut on and cut off of the band pass Butterworth filter. Higher value makes the filter steeper. |
| 6 | Python.exe in the Cellpose Environment | Text | *.exe file, without arguments |
| 7 | Cellpose Model | Text | Model for Cellpose cell segmentation. This model should support additional nuclear marker. |
| 8 | Cellpose Diamenter (pixels) | Text | Approximate diameter of cells |
| 9 | Cellpose --mask_threshold | Text | Maximum allowed error of the flows for each mask. |
| 10 | Cellpose --flow_threshold | Text | Minimum probability to return cell segments |
| 11 | Cellpose Number of parallel processes | Integer | Image series and time courses will be split into multiple parts and separate instances of the external program is launched. the external program is launched. |
| 12 | Cellpose Batch Size | Text | Set batch size according to the GPU memory |
Structure
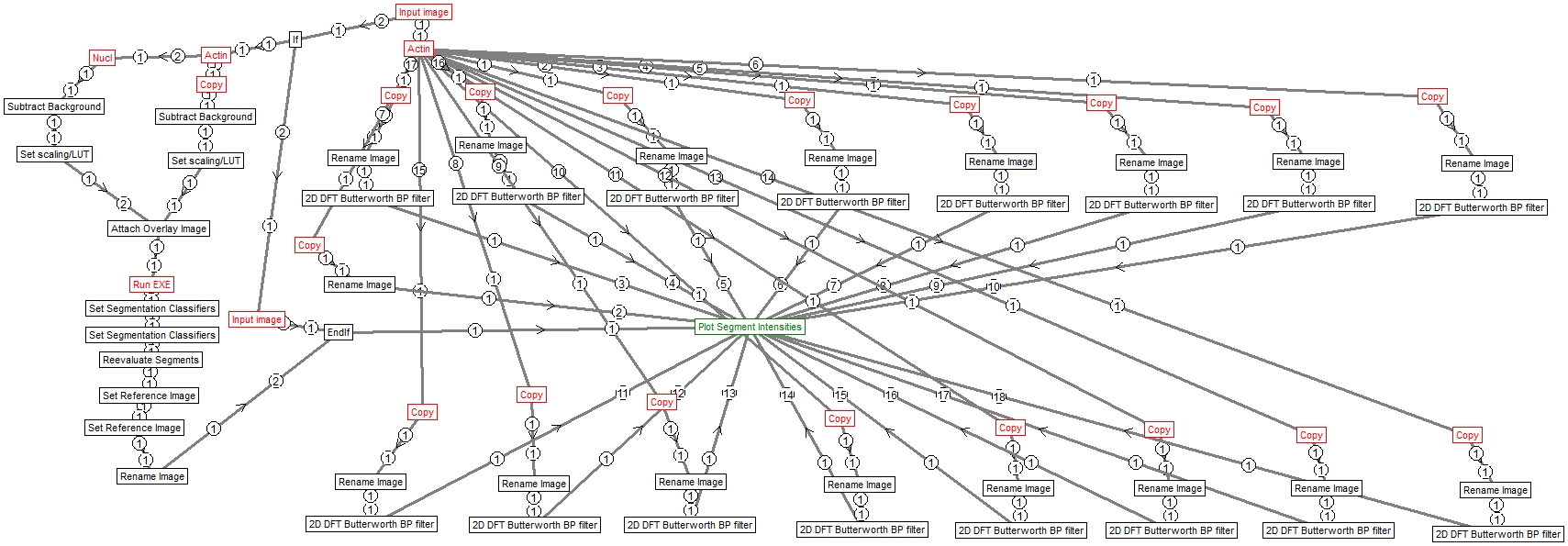
Image Analyst MKII pipeline – saved by V4.2.5 (build 838)